The primary function of the CMIC Structural Biology Core is to facilitate high-quality sample preparation for use in our X-ray crystallography as well as single particle reconstruction in cryo-electron microscopy.
How to access:
All users who want to use CMIC Structural Biology core’s X-ray crystallography and/or Cryo-EM pipelines need to contact Dr. Eric Enemark (BioMed I, Rm 405). CMIC SB core uses Outlook Calendar System for Instruments booking. Please contact Dr. Eric Enemark for authorization. Users needing assistance or training before use of instruments need to contact Dr. Rahul Jaiswal (BioMed I, Rm 406).
Services
Sample preparation: protein design, expression, purification, and quality control
A major function of the SBC is to help facilitate high-quality sample preparation for projects involving structural biology or biophysical studies, mostly focused on protein production. State-of-the-art equipment for protein expression in bacterial, insect, and mammalian expression hosts is available to core users.
X-ray crystallography
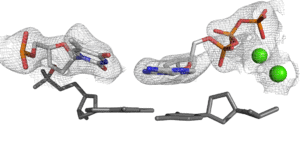
For X-ray crystallography, SBC helps with crystal screening, optimization, synchrotron data collection, data analysis and structure solution. The general pipeline is to first perform crystallization screens with a Mosquito drop-setting robot using standard commercially available kits. Crystallization plates are automatically monitored in a Formulatrix RockImager-1000 plate incubator/hotel. Lead crystallization conditions are optimized by standard methods, such as crystallization grid screens and seeding. Suitable crystals are cryoprotected, flash-frozen, and sent to the SER-CAT facility at APS. Data are collected by remote operation of SER-CAT beamlines. X-ray crystal structures are solved and refined using standard software on Linux workstations.
Cryo-Electron microscopy: Single Particle & Cryo-ET

For single particle cryo-EM, SBC assists with sample preparation, grid preparation (negative staining and cryo-EM), grid optimization, collection, processing, and data analysis. The general pipeline is to first analyze samples by negative stain EM using the TF20 electron microscope maintained by the UAMS Digital Microscopy Core. The Structural Biology Core will pay internal instrument charges to access this microscope. Following suitable negative stain screening, an application is initiated to the NIH Transformative High-Resolution Cryo-electron Microscopy program, and samples are frozen to grids with a Vitrobot. Frozen grids can be screened with the TF20 microscope in the UAMS Digital Microscopy Core. The NIH Cryo-EM program has programs with preferred access for IDeA states, and UAMS currently uses an EM grid-clipping system from the Pacific Northwest Center for Cryo-EM within this program. After a sample is allocated time at an NIH center, frozen grids are shipped to the NIH facility, and data are collected by support staff of the NIH center. The resulting data are downloaded to a UAMS storage server in the Structural Biology Core for processing, structure determination, and refinement using a 4-GPU workstation. The Structural Biology Core has budgeted to purchase a 4-GPU workstation dedicated to RPL projects. The Enemark lab also has two 4-GPU workstations, and these computational resources will be available to Core projects when not active on projects of the Enemark lab. Access to cryo-electron tomography techniques will use an analogous pipeline to obtain instrument time and suitable training in the NIH National Network for Cryo-ET. The Structural Biology Core has budgeted funds to purchase microscope time for high-resolution data collection for any cases where instrument time is not obtained in a timely way from the NIH Cryo-EM program.
More Information
For more information or to set up a consultation to discuss research goals, please contact:

How to Acknowledge the CMIC
“Research reported in this [publication, release] was supported (in whole or in part) by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P20GM152281. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
For abstracts, a short note “Supported by a grant from the NIH/NIGMS – 1P20GM152281” will suffice.